A Chemical Biology Approach for de novo Discovery of Selectively Essential Metabolic Enzymes
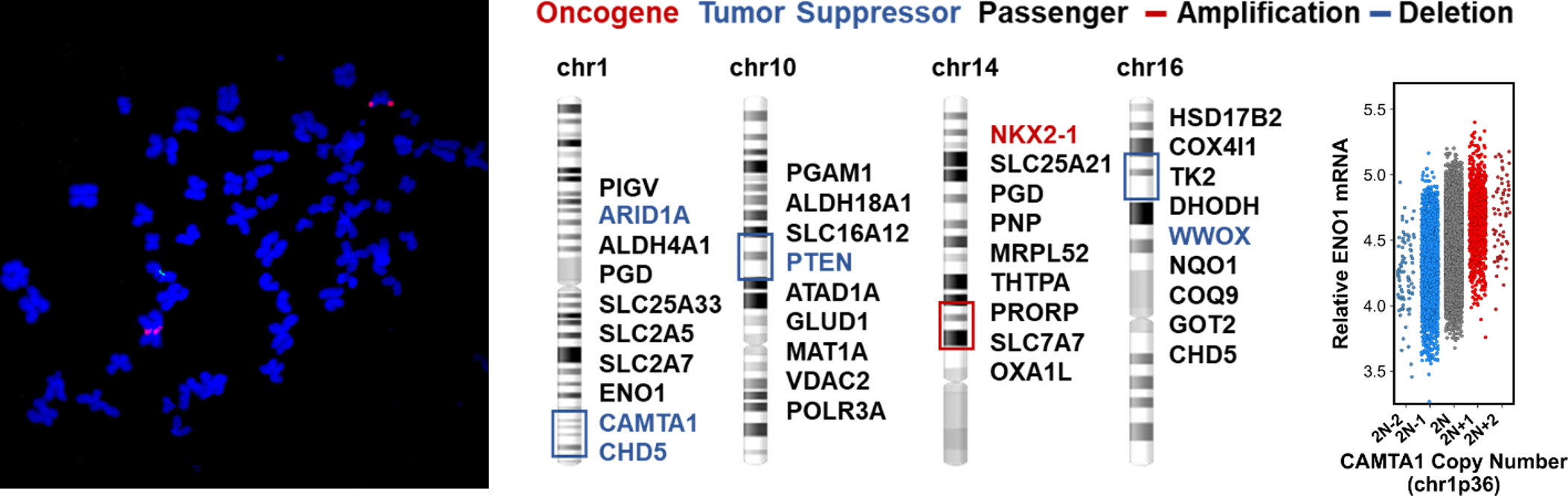
In addition to gain- and loss-of-function mutations in oncogenes and tumor suppressors, copy number alterations (CNAs) that significantly affect the expression of these proteins play a pivotal role in tumorigenesis. While oncogenes and tumor suppressors are often challenging to target with small molecules, CNAs collaterally impact the expression of thousands of metabolic enzymes, which present promising opportunities for therapeutic intervention. The Hanigan lab focuses on investigating a specific subset of these enzymes, particularly those associated with the recessive metabolic disorders known as inborn errors of metabolism (IEM), with the goal of developing selective therapeutics for cancer and IEM.
Figure 1: Collateral copy gain/loss of recessive disease-causing enzymes results in actionable cancer vulnerabilities
Catabolic enzymes as drug targets
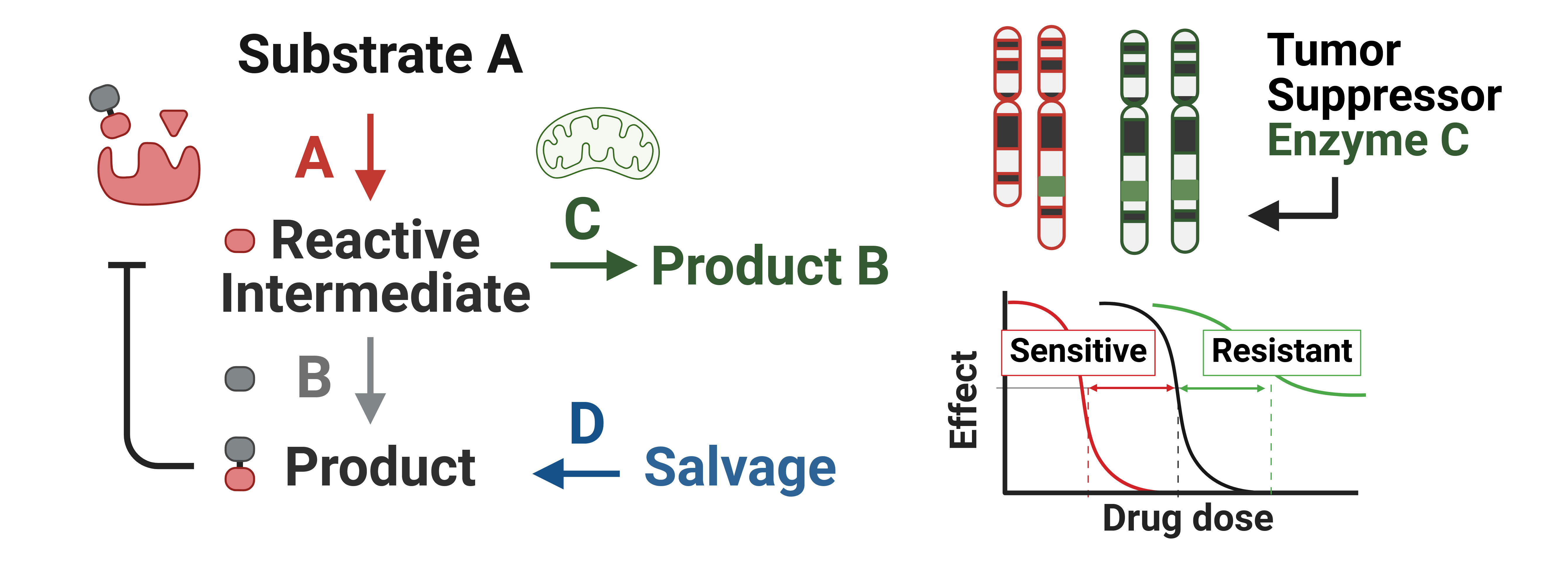
In general, IEM result from biallic loss-of-function mutations in catabolic enzymes that break down reactive electrophilic or nucleophilic intermediary metabolites, which accumulate and spontaneously react with essential cellular proteins, nucleic acids, or cofactors. Thus, these enzymes represent “validated” targets to specifically induce cell type specific toxicity through drug induced haploinsufficiency in cancer cells where the target enzyme has been collaterally deleted. Moreover, as opposed to biosynthetic enzymes, which often have redundant mechanisms to salvage or produce metabolic endproducts required for cell proliferation, targeting catabolic enzymes should be less prone to drug resistance through metaoblic plasticity.
Figure 2: Metabolic pathway showing a vulnerabiilty resuting from hemizygous deletion of a catabolic enzyme (green).
Leveraging metabolism to identify and drug metabolic vulnerabilities
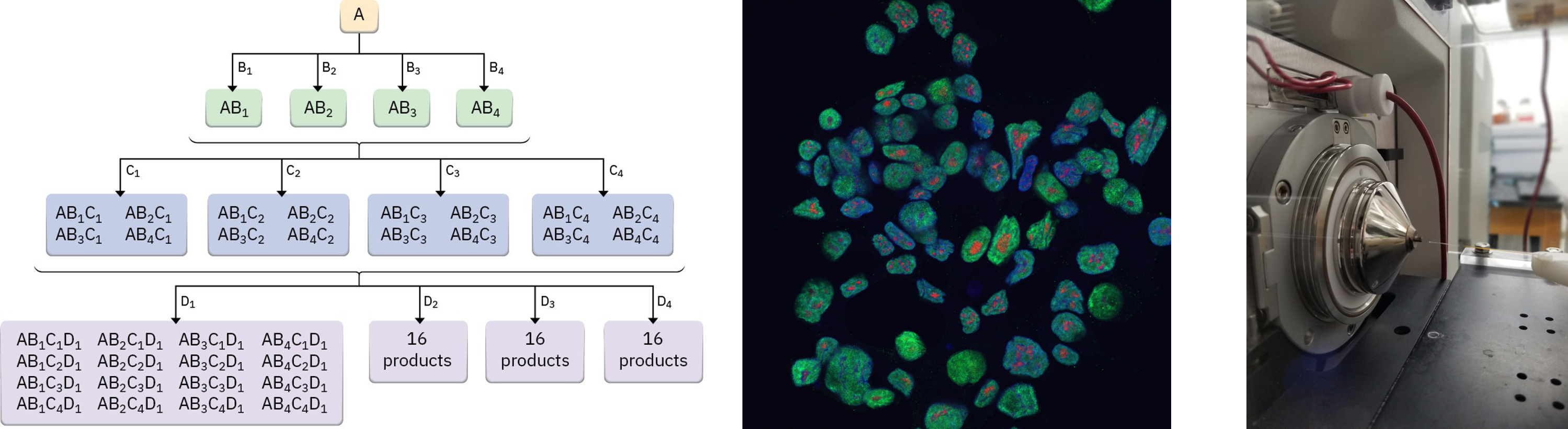
To accomplish these goals, we use an interdisciplinary approach combining synthetic organic chemistry, chemical biology and systems pharmacology. Lab projects are focused on developing combinatorial synthetic platforms that leverage native metabolic reactions, methods to generate cellular models of recurrent CNA in cancer and metabolic disease, and novel chemical-proteomic and -genetic approaches to identify therapeutic targets of interest. These projects combine both method development and application of these methods for basic and translational biology.
Figure 3: Combinatorial chemistry, high-content imaging and chemical proteomics.
.png)